Scroll to:
Composition and dynamics of bladder, vaginal and bowel microbiota during three trimesters in healthy pregnant women
https://doi.org/10.21886/2308-6424-2024-12-5-12-23
Abstract
Introduction. The microbiota dynamics of the core biotopes during pregnancy are hardly studied, although changes in these compartments have an important role in both the functioning of the female organism and foetal development.
Objective. To study the dynamics and interactions of changes in bladder, vaginal and bowel microbiota in healthy pregnant women over three trimesters (TRI-1, 2, 3).
Materials & Methods. Study design: a single-centre comparative observational longitudinal study. Thirty out of first-time 220 pregnant women were selected for screening at the antenatal clinic from 2021 to 2022. All pregnant women underwent sampling at T-1, 2, 3: mid-stream bladder urine samples, posterior vaginal swabs and faecal masses were collected for culture study. After a special pre-culture preparation, samples were examined on an expanded set of nutrient media (n = 13) using special cultivation (aerobic-anaerobic) conditions. Based on the research results, identification frequencies (IDFs), microbial load values (MLVs) and microbial co-occurrence coefficients between the different biotopes were estimated.
Results. Culture study revealed various bacteria in each biotope investigated during all TRIs. In the urine, aerobes and anaerobes were observed from TRI-1 to TRI-3 with different IDFs, but no taxa showed a stable IDFs. In the vagina, IDFs of bacteria were similar to urinary. The bowel microbiota was the most stable biotope remained almost unchanged during pregnancy. In the urine and vagina, mean MLVs of most aerobes and anaerobes did not change significantly throughout pregnancy. In the bowel, MLVs were consistently higher than in the urine and vaginal swabs. According to the co-occurrence analysis bladder-vagina and bladder-bowel biotopes showed significantly more interconnections between microorganisms in all TRIs.
Conclusion. The observed microbiota structure during all TRIs is associated with uncomplicated gestation. These results will be valuable for studying changes of microbiota in complicated pregnancies.
For citations:
Naboka Yu.L., Vorobyeva N.V., Gudima I.A., Sinyavskaya T.G., Ismailov R.S., Naber K.G., Kogan M.I. Composition and dynamics of bladder, vaginal and bowel microbiota during three trimesters in healthy pregnant women. Urology Herald. 2024;12(5):12-23. https://doi.org/10.21886/2308-6424-2024-12-5-12-23
Introduction
Pregnancy involves a series of metabolic changes in the female physiology, primarily in the hormonal and immune systems. Recent studies have identified important rearrangements in the bowel, vaginal, urinary, and oral microbiota and microbiome in pregnant women playing a definite role in pregnancy course and complications [1][2].
There are plenty of microorganisms, i.e. bacteria, viruses, and fungi, that have an impact on human host health. Undoubtedly, the maternal microbiota has a predominant influence on the formation, maturation, and development of microbial communities in foetal and neonatal organs and systems. From this perspective, it is essential to have an insight regarding the microbiota in key biotopes of healthy pregnant women (HPW) during first, second and third trimesters (TRI-1, TRI-2, TRI-3) [2][3].
By now it is known, the TRI-1 bowel microbiota resembles that of healthy non-pregnant women. However, evidence for changes in the bowel microbiota from TRI-1 up to delivery is limited [3] and strongly associated with individual heterogeneity [1][3].
It has been determined that bowel microbiota shift during pregnancy aims to cover foetal energy and metabolic needs [1][4]. Diet during TRI-1 and TRI-2 affects the bowel microbiota composition in the TRI-3 [5], and the formation of neonatal microbiota [6]. Several studies indicate that bowel microbiota shift is associated with an increased risk of gestational arterial hypertension, diabetes mellitus, obesity, early preeclampsia, and pregnancy loss [7 – 12].
It is well recognised how the vaginal microbiota changes throughout a female lifecycle and also related to their menstrual calendar [13]. But it is now also clear that vaginal dysbiosis in pregnancy increases the risk of adverse obstetric outcomes, including spontaneous preterm delivery [14 – 16]. In a healthy woman, the lactobacilli that dominate the vaginal discharge contribute to maintaining a low pH, which protects this biotope from bacterial and fungal infections [17]. In turn, altered proportions between lactobacilli and other aerobes/anaerobes in the vaginal microbiota of HPW can occur, increasing the risk of dysbiosis and elevating the risk of dysbiosis and premature delivery chance [18]. There are also changes in lactobacilli composition, so the most found L. crispatus in European women is replaced by L. iners [18], which is specifically associated with premature pregnancy loss [19][20]. In HPW, some studies have shown that the TRI-3 vaginal microbiota is analogous to the pre-pregnancy microbiota, but significant changes occur in the vaginal microbiota after childbirth [21]. On contrary, several studies have refuted the resemblance of the vaginal microbiota during pregnancy with that of non-pregnant women, citing both the low taxa diversity and the absence or presence of unique bacteria [22][23].
Five types of vaginal microbiome in asymptomatic non-pregnant North American women [24][25], along with differences in the microbiome of white and black women and its ethnicity patterns during pregnancy have been established in several studies [26 – 30]. Simultaneously, there are very limited concurrent studies available on the vaginal and bowel microbiota in pregnant women [31 – 33]. High interindividual variability and dynamic composition of microbial communities depending on gestational age in physique biotopes have been demonstrated [32]. Bowel lactobacilli decline sharply after TRI-1 while vaginal lactobacilli are stable throughout the three TRIs [33]. Regarding research on the bladder microbiota during an uncomplicated pregnancy in HPW, these studies are extremely valuable in assessing the so-called asymptomatic bacteriuria. Researchers consider it a high-risk factor for gestational pyelonephritis, maternal sepsis, and preterm labour [34][35]. Therefore, screening for asymptomatic bacteriuria and its treatment during pregnancy is recommended [36 – 39]. But is it right way?
In the last decade, the belief that the urinary system is sterile has been challenged [40][41]. The bladder urine microbiota in healthy women has been studied quite fully throughout their lives [42 – 46].
The bladder urine microbiota in HPW throughout all trimesters has also been studied, and a diverse range of urinary aerobes and anaerobes has been identified. However, no comprehensive study of the three biotopes (bladder, vagina, bowel) has been conducted in HPW to date.
Objective. To study the dynamics and interactions of changes in bladder, vaginal and bowel microbiota in healthy pregnant women over three trimesters (TRI-1, 2, 3).
Materials and methods
1. Ethical statement
The study was registered by the Ethics Committee of Rostov State Medical University (Protocol №15/20 dated 08/10/2020) [47]. Study design: a single-centre comparative observational longitudinal study.
2. Patient brief demographics
From 2021 to 2022, 30 HPW were sequentially included according to criteria planned (Table 1) in a single-center prospective cohort observational study out of 280 who applied to antenatal clinics for the diagnosis of pregnancy during the TRI-1. All participants have read and signed a voluntary informed consent.
Demographic data, sexual life patterns, urogenital symptoms were registered for all pregnant women.
Table 1. Criteria for selection of healthy pregnant women
|
Inclusion criteria |
Exclusion criteria |
|
• healthy pregnant women aged 20 – 35 years; • singleton primary pregnancy verified before 12 weeks; • absence of any comorbidities; • history of acute lower urinary tract infections (UTI) but no neurogenic lower urinary tract symptoms or infection signs within 3 months prior to pregnancy. |
• pregnancy due to assisted reproductive technologies; • kidney and urinary tract anomalies and diseases; • vaginal dysbiosis; • use of antibiotics since conception; • history, and current presence of sexually transmitted infections, • HIV positive; • drug addiction & abuse; • smoking. |
3. Diagnostic work-up
Pregnant women underwent obstetric examination at gestational ages of 8 – 12 weeks (TRI-1), 22 – 24 weeks (TRI-2) and 32 – 36 weeks (TRI-3). Midstream urine samples, posterior vaginal discharge and faecal masses were collected from all pregnant women for culture study at times under research.
All women completed their pregnancies at 37 to 38 weeks by term vaginal labour (22.0 – 73.3%) or C-section (8.0 – 26.7%) and delivered healthy neonates.
- Sampling
Midstream urine samples were collected by trained medical staff after appropriate hygienic procedure when pregnant women urinated independently. Urine was collected in a sterile disposable container (Sterile UricolTM — “HiMedia Laboratories Pvt. Ltd.”, Mumbai, India).
Vaginal samples were collected using a disposable gynaecological mirror and pH was measured using pH strips (colpo-pH test). Vaginal discharge was collected using two sterile swabs with unique identification numbers. One swab was used to prepare a smear followed by Gram staining (Gram Stains-Kit — “HiMedia Laboratories Pvt. Ltd.”, Mumbai, India) to assess the vaginal microbiota according to the Nugent R.P. (1 – 10 scale) [48]. Other swabs with transport medium (Hiculture Transport Swabs w/Alternative Thioglycollate Medium — “HiMedia Laboratories Pvt. Ltd.”, Mumbai, India) were used for culture study.
Faecal samples were collected in a sterile container (Sterile ClinicolTM — “HiMedia Laboratories Pvt. Ltd.”, Mumbai, India) with a unique identification number.
- Culture study
Urine culture was performed by streaking to quantitatively count the grown colonies on suitable nutrient media [49]. Tampons with vaginal discharge were placed in a tube with 1.0 mL of 0.9% PBS and gently shaken. Further serial dilutions were performed with transfer of 100 µl each into the next tubes with 0.9% PBS. By this way dilutions up to 107 were obtained [33]. From each dilution, seeding (100 µl) were performed on appropriate nutrient media to count the quantity of microorganisms.
Faeces (1000 mg) were homogenised in 9.0 mL of pre-reduced PBS. Subsequently, nine-fold dilutions were performed, and each sample was cultured on the appropriate nutrient media to count the quantity of microorganisms [50].
In our study we used an advanced nutrient media set for the cultivation of aerobes and anaerobes: MacConkey Agar, HiCrome Klebsiella Selective Agar Base, HiCrome Candida Differential Agar, HiCrome Enterococci Agar, HiCrome Aureus Agar Base, Blood Agar Base, Streptococcus Selection Agar, Rogosa SL Agar, Bifidobacterium Agar, MRS Agar, Anaerobic Agar, Shaedler Agar, Bacteroides Bile Esculinum Agar (“HiMedia Laboratories Pvt. Ltd.”, Mumbai, India). The samples were cultivated under aerobic (24 – 48 h) and anaerobic (48 – 72 h) conditions using a AnaeroHiGas Pak (“HiMedia Laboratories Pvt. Ltd.”, Mumbai, India). Microorganisms were identified based on their morphological and haemolytic properties. Smears were prepared from colonies grown in culture medium and heat-fixed for Gram staining according to standard protocol using an oil-immersion microscope (magn. х 900, 10 – 15 FoV). Final identification of microorganisms was carried out according to their biochemical properties using entero-, staphylo-, and anaerotests (“Erba Lachema s.r.o.”, Brno, Czech Republic).
4. Statistics methods
The statistical software IBM SPSS Statistics 26.0 («SPSS: An IBM Company», IBM SPSS Corp., Armonk, NY, USA) was used to perform the calculations. The microbial identification frequency (IDF, %) and average microbial load values (MLV, CFU/mL) in the biotopes of HPW were calculated. The mean values permitted the inclusion of low-frequency high levels of MLVs and were used for comparative characterisation rather than as a central measure of distribution. The Mann-Whitney U test was used to compare IDFs and MLVs. Relationships between microbes in different biotopes was investigated using Pearson's coefficient of reciprocal conjugation (co-occurrence coefficient). Standard statistical significance levels of 1% and 5% were used for hypothesis testing.
Results
In HPW, culture study during TRI-1, TRI-2, and TRI-3 revealed various aerobes and anaerobes in each biotope investigated. IDFs of selected bacteria throughout the three TRIs are shown in Figure 1 and Figure 2.
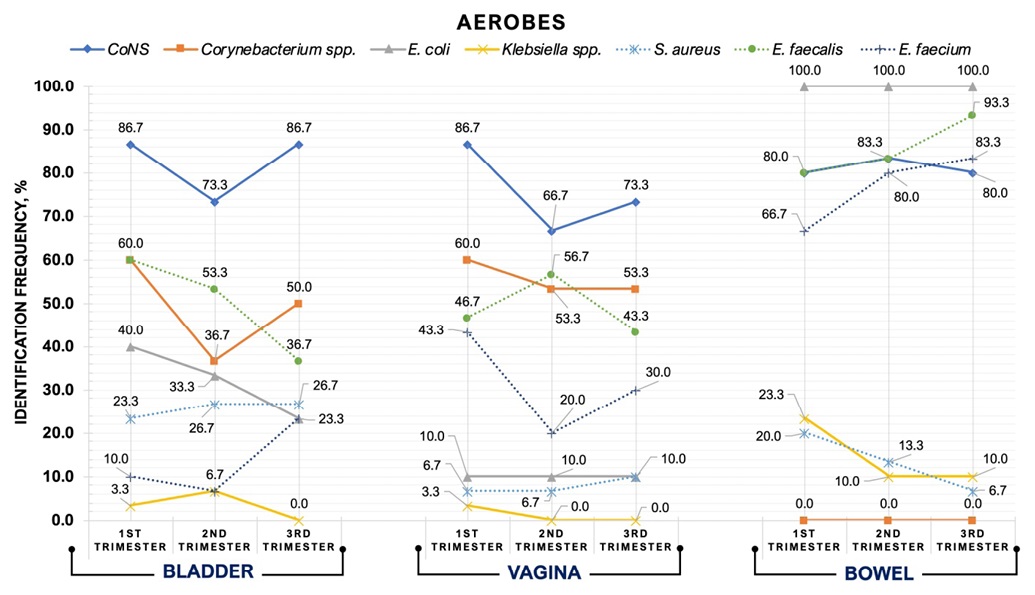
Figure 1. Identification frequency trends of aerobes in three biotopes during pregnancy
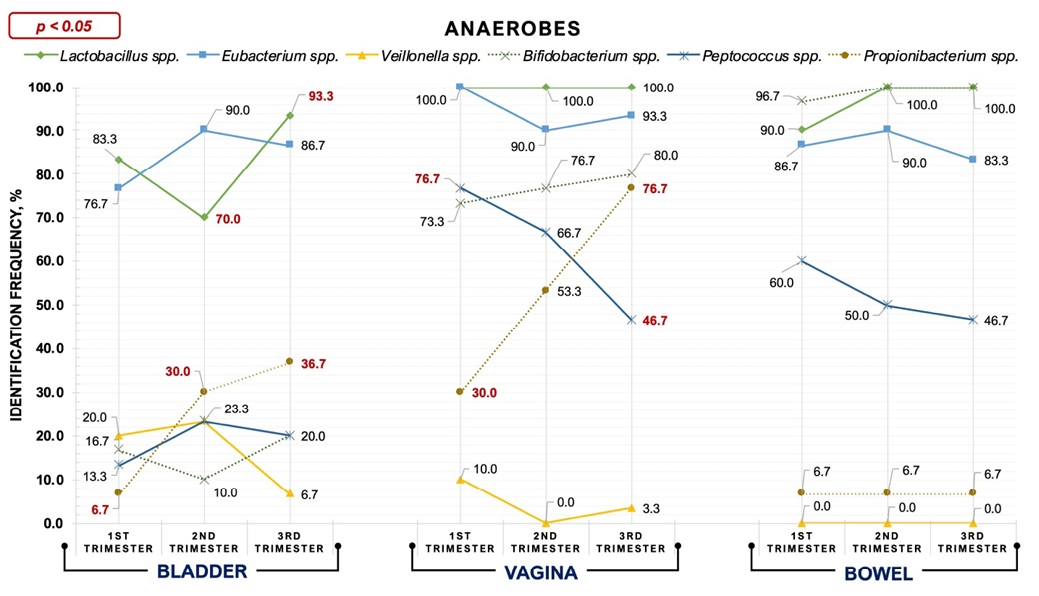
Figure 2. Identification frequency trends of anaerobes in three biotopes during pregnancy
1. Evaluation of identification frequency
- Bladder urine microbiota
In bladder urine samples, aerobes and anaerobes were detected from TRI-1 to TRI-3 with different IDFs. However, multidirectional trends were found in the urine microbiota regarding IDFs. Thus, IDFs for CoNS, Corynebacterium spp., E. faecium, Lactobacillus spp. and Bifidobacterium spp. decreased during TRI-2. In contrast, detection rates for Klebsiella spp., Eubacterium spp., Peptococcus spp., and Veillonella spp. were highest throughout TRI-2. Only two taxa (E. faecalis, E. coli) showed a decrease in IDFs from TRI-1 to TRI-3. Generally, E. faecium, Lactobacillus spp. (TRI-1 vs TRI-3, p < 0.05), Propionibacterium spp. (TRI-1 vs TRI-3, p < 0.05) and Bifidobacterium spp. had the highest urinary detection rates during TRI-3. Only Klebsiella spp. Were eliminated from the urine during TRI-3.
- Vaginal microbiota
Vaginal bacteria detection frequency trends were similar to those for urine. Stable vaginal inhabitants of HPW throughout all TRIs were Lactobacillus spp. (100.0%), and E. coli isolated with markedly reduced frequency (10.0%). By TRI-3, five aerobes and three anaerobes demonstrated decreasing IDFs. In contrast, S. aureus, Bifidobacterium spp. and Propionibacterium spp. marked an increase in MLV (TRI-1 vs TRI-3, p < 0.05). Klebsiella spp were eliminated from vaginal secretions during TRI-2 and TRI-3, and Veillonella spp. exceptionally during TRI-2.
- Bowel microbiota
Bowel microbiota showed to be the most stable biotope. Thus, E. coli, Propionibacterium spp. and CoNS remained essentially unchanged in their IDFs. E. faecalis, E. faecium, Lactobacillus spp. revealed the baseline high IDFs during TRI-1 increased throughout TRI-3, but Eubacterium spp. and Peptococcus spp. revealed a decrease in IDFs. Klebsiella spp. and S. aureus also diminished towards the TRI-3. Corynebacterium spp. and Veillonella spp. were not detected in the bowel microbiota throughout pregnancy.
2. Evaluation of microbial load values
The average MLVs of bacteria in the studied biotopes are shown in Figure 3 and Figure 4.
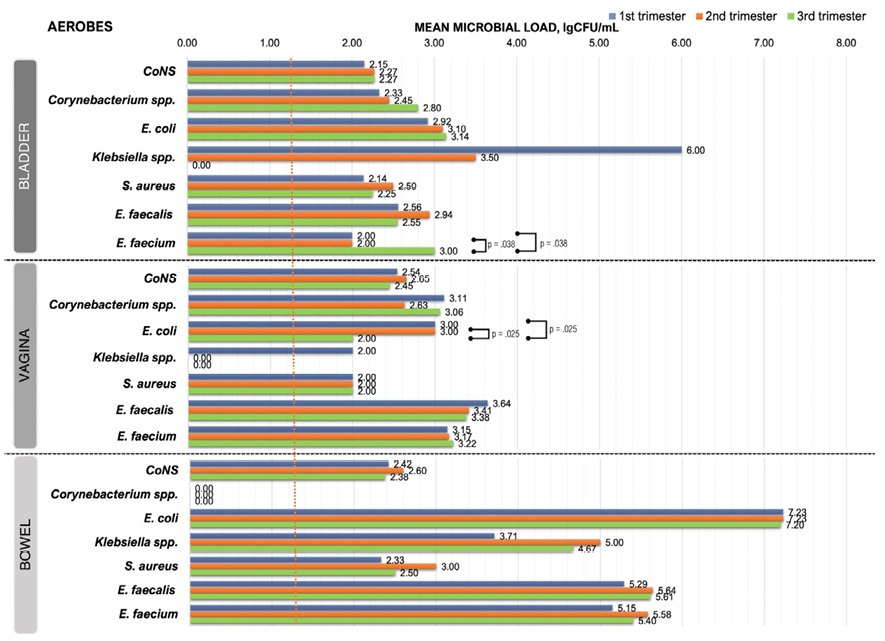
Figure 3. Microbial load dynamics of aerobes from three biotopes during pregnancy (red dotted line — detection limit, 0.00 — below detection limit
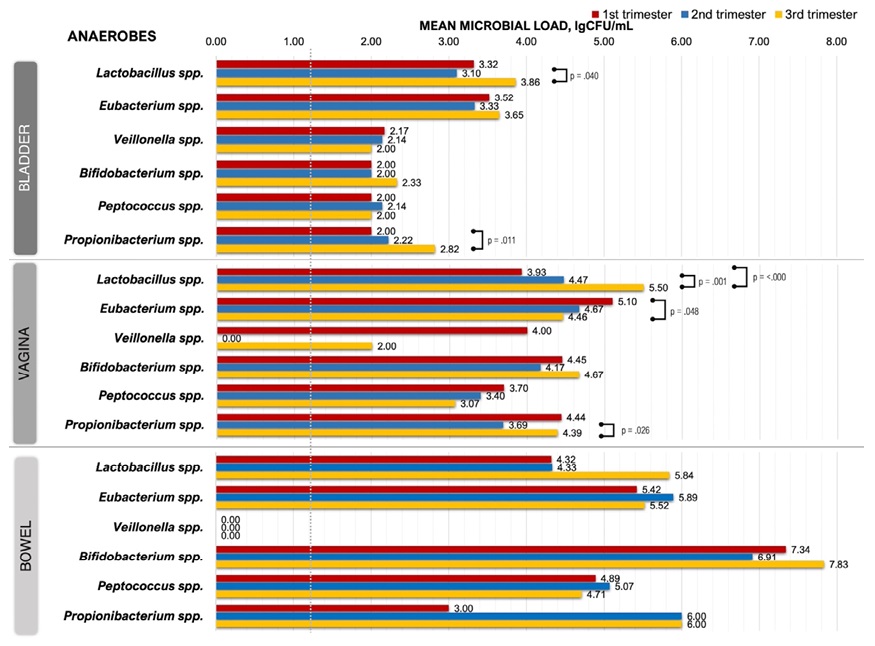
Figure 4. Microbial load dynamics of anaerobes from three biotopes during pregnancy (green dotted line — detection limit, 0.00 — below detection limit)
- Bladder microbiota
During TRI-1, mean baseline MLVs for most bacteria were < 103 CFU/ml. Only Klebsiella spp., Lactobacillus spp. and Eubacterium spp. had mean bacteriuria levels of 106.00, 103.32 and 103.52 CFU/ml respectively.
By TRI-3, mean MLV did not change significantly for most aerobes and anaerobes, only E. faecium (TRI-1 vs TRI-3, p = 0.038; TRI-2 vs TRI-3, p = 0.038), Lactobacillus spp. (TRI-2 vs TRI-3, p = 0.040) and Propionibacterium spp. (TRI-1 vs TRI-3, p = 0.011) showed a significant increase in bacteriuria. Among aerobes, only Klebsiella spp. was non-trend-specific, having a MLV of 106.00 CFU/ml in the T1 that completely reduced through the TRI-2 to zero-level in TRI-3.
- Vaginal microbiota
Most vaginal aerobes showed that initial (TRI-1) MLVs slightly higher than urinary, excluding Klebsiella spp. and S. aureus. Fluctuations in aerobic MLVs were insignificant during pregnancy. The differences in MLVs between TRI-1 and TRI-3 were significant only for E. coli (TRI-1 vs TRI-3, p = 0.025; TRI-1 vs TRI-3, p = 0.025).
In TRI-1, vaginal anaerobes showed MLV generally higher than urinary. The overall MLV of vaginal discharge throughout pregnancy was elevated compared to urinary. Lactobacillus spp. showed a significant MLV increase from TRI-1 to TRI-3 (TRI-1 vs TRI-3, p < 0.001; TRI-2 vs TRI-3, p = 0.001), and Eubacterium spp. demonstrated its decrease (TRI-1 vs TRI-3, p = 0.011).
- Bowel microbiota
Bowel microbiota was characterized by consistently high MLVs (>10⁵ CFU/ml), specified for E. coli, E. faecalis, E. faecium, Eubacterium spp., Bifidobacterium spp. и Propionibacterium spp. However, it should be noted that deviations of MLV for faecal samples with aerobes and anaerobes were unreliable between TRIs.
3. Evaluation of biotope interactions
Indices of co-occurrence (contiguity) coefficient between taxa of the three biotopes based on verification frequency during TRIs are presented consecutively in Tables 2, 3, and 4.
Table 2. Co-occurrence coefficients for identification frequencies between taxa of the three biotopes in first trimester
|
Microorganisms |
Urine — Vagina |
Urine — Bowel |
Vagina — Bowel |
|||
|
COC* |
Р |
COC* |
Р |
COC* |
Р |
|
|
Corynebacterium spp. |
0.504 |
0.001** |
– |
– |
– |
– |
|
CoNS |
0.356 |
0.037*** |
0.426 |
0.01*** |
0.387 |
0.25 |
|
S. haemolyticus |
0.431 |
0.488 |
0.147 |
0.414 |
0.378 |
0.025** |
|
S. saprophyticus |
0.659 |
< 0.001*** |
0.513 |
0.001*** |
0.563 |
< 0.001*** |
|
S. lentus |
0.484 |
0.002*** |
0.545 |
< 0.001*** |
0.463 |
0.004*** |
|
S. warneri |
0.63 |
< 0.001*** |
0.579 |
< 0.001*** |
0.523 |
0.001*** |
|
S. epidermidis |
0.392 |
0.02** |
0.522 |
0.001*** |
0.554 |
< 0.001*** |
|
Enterococcus spp. |
0.537 |
< 0.001*** |
0.14 |
0.439 |
0.13 |
0.472 |
|
E. faecalis |
0.531 |
0.001*** |
0.263 |
0.136 |
0.288 |
0.1 |
Note. CoNS — coagulase-negative staphylococci; *COC — co-occurrence coefficient; ** — differences are significant at 5% level; *** — at 1% level; "–" — coefficients are not calculated if values are constants
Table 3. Co-occurrence coefficients for identification frequencies between taxa of the three biotopes in second trimester
|
Microorganisms |
Urine — Vagina |
Urine — Bowel |
Vagina — Bowel |
|||
|
COC* |
Р |
COC* |
Р |
COC* |
Р |
|
|
CoNS |
0.35 |
0.041** |
0.596 |
< 0.001*** |
0.245 |
0.166 |
|
S. haemolyticus |
0.311 |
0.043** |
0.323 |
0.061 |
0.258 |
0.144 |
|
S. saprophyticus |
0.336 |
0.051 |
0.533 |
0.001*** |
0.336 |
0.051 |
|
S. lentus |
0.471 |
0.351 |
0.617 |
< 0.001*** |
0.563 |
< 0.001*** |
|
S. warneri |
0.555 |
0.467 |
0.362 |
0.033** |
0.251 |
0.156 |
|
S. epidermidis |
0.542 |
0.584 |
0.442 |
0.007*** |
0.441 |
0.007*** |
|
E. faecalis |
0.252 |
0.031** |
0.119 |
0.513 |
0.149 |
0.41 |
|
Eubacterium spp. |
0.111 |
0.042** |
0.251 |
0.156 |
0.11 |
0.543 |
|
Peptococcus spp. |
0.363 |
0.433 |
0.079 |
0.666 |
0.272 |
0.121 |
|
Fusobacterium spp. |
0.707 |
< 0.001*** |
0.707 |
< 0.001*** |
0.707 |
< 0.001*** |
Note. CoNS — coagulase-negative staphylococci; *COC — co-occurrence coefficient; ** — differences are significant at 5% level; *** — at 1% level
Table 4. Co-occurrence coefficients for identification frequencies between taxa of the three biotopes in third trimester.
|
Microorganisms |
Urine — Vagina |
Urine — Bowel |
Vagina — Bowel |
|||
|
COC* |
Р |
COC* |
Р |
COC* |
Р |
|
|
Corynebacterium spp. |
0.372 |
0.028** |
0.183 |
0.309 |
0.195 |
0.277 |
|
S. haemolyticus |
0.504 |
0.001*** |
0.471 |
0.003*** |
0.471 |
0.003*** |
|
S. saprophyticus |
0.626 |
< 0.001*** |
0.336 |
0.051 |
0.071 |
0.696 |
|
S. lentus |
0.15 |
0.935 |
0.607 |
< 0.001*** |
0.617 |
< 0.001*** |
|
S. warneri |
0.383 |
0.087 |
0.513 |
0.001*** |
0.571 |
< 0.001*** |
|
S. epidermidis |
0.289 |
0.099 |
0.36 |
0.035** |
0.067 |
0.713 |
|
Eubacterium spp. |
0.253 |
0.041** |
0.523 |
0.001*** |
0.119 |
0.513 |
|
Peptococcus spp. |
0.281 |
0.651 |
0.345 |
0.044** |
0.193 |
0.282 |
|
Peptostreptococcus spp. |
– |
– |
– |
– |
0.707 |
< 0.001*** |
Note. CoNS — coagulase-negative staphylococci; *COC — co-occurrence coefficient; ** — differences are significant at 5% level; *** — at 1% level; «–» — coefficients are not calculated if values are constants
As observed in TRI-1, high-significant association strengths were found between microorganisms verified in biotopes.
In TRI-2, reduction of such high-significant association strengths characterising the tightness of taxa interrelationships in biotopes was observed. In TRI-3, the probability of such associations becomes higher again.
Bladder and bowel biotopes were more strongly associated in all TRIs, but such associations between vaginal and bowel biotopes occurred less in TRI-2 and TRI-3. Notably, the sought co-occurrence coefficients were highly significant in all TRIs and were greatly marked for aerobes, specifically for CoNS cluster species. By contrast, the estimated coefficients for anaerobes between biotopes were observed only for certain taxa.
Discussion
Pregnancy is a period of profound changes in the female physique, both to adapt for the new conditions of functioning and to support and develop the foetus. The female microbiota plays a fundamental role in these processes, although insights into the structural and qualitative microbiota features of different physique sites of pregnant women are still clearly limited. This applies both to cross-biotopic relationships and their impact on the course of healthy and complicated pregnancies. The vaginal microbiota has been studied better than other biotopes in non-pregnant and pregnant women [23][24][30]. Studies have shown greater stability of the vaginal microbiota during pregnancy than in the non-pregnant state [23] and the impact on its variation from gestational age [1]. In fact, it has been observed that pregnancy progression does not result in significant taxonomic changes in the vaginal microbiota [31].
The results of our analysis based on microbiota evaluation of exclusively HPW over three TRIs, reveal precisely reliable changes in detection frequencies of most taxa from TRI to TRI, while the IDF of only Lactobacillus spp. and E. coli remained stable. Furthermore, no significant changes were observed in the IDFs of vaginal aerobes and anaerobes during pregnancy. However, TRI-3 showed a remarkable increase in MLV for Lactobacillus spp., which was accompanied by a significant decrease in MLVs by E. coli and Eubacterium spp.
Previous studies of the bowel microbiota have revealed an increasing diversity of taxa from TRI-1 to TRI-3 [1]. Conversely, our study demonstrated just stability for taxa of this biotope both in IDFs and MLVs. But the variations of some microorganisms in the upward or downward trend by TRI-3 turned out to be insignificant. In the last 15 years, no studies have investigated the bladder urine of pregnant women along with vaginal and bowel biotopes, although isolated studies on urine in pregnant women have certainly been conducted [34][35].
None of the 25 urinary microorganisms studied showed stability in IDFs and MLVs. Trends for IDFs of urinary microbes from TRI-1 to TRI-3 were multidirectional. The greatest changes were seen in most bacteria during TRI-2. This was especially true for anaerobes. The average bacteriuria for most aerobes and anaerobes did not exceed 103 CFU/ml. However, some anaerobes exhibited a significant increase in bacteriuria by TRI-3. Among the all aerobes, only Klebsiella spp. showed high bacteriuria values (10⁶ CFU/mL) in TRI-1 and no bacteriuria at all during TRI-3.
In bladder and vaginal microbiota, CoNS, Corynebacterium spp., E. coli, and E. faecalis were dominant aerobes, while Lactobacillus spp., Eubacterium spp. were dominant among the anaerobes. In addition, Bifidobacterium spp., Propionibacterium spp., and Peptococcus spp. were often found in vaginal discharge. In bowel microbiota, E. coli was a common microorganism, as are E. faecalis and E. faecium, and CoNS were also high-frequency prevalent in all TRIs The dominant anaerobic bacteria in this biotope included Bifidobacterium spp., Lactobacillus spp., and Eubacterium spp.
Thus, we determined that CoNS and Lactobacillus spp were the dominant in biotopes at all TRIs. Our data on the IDFs of Lactobacillus spp in biotopes are not complementary to the results of other studies, especially regarding to the bowel Lactobacillus spp [31][32]. The differences may be related to the heterogeneity of the pregnant women cohort where premature births occurred, with the specific dietary and nutritional habits, use of progesterone treatments [52], as well as with the predominant use of sequencing techniques to identify bacteria in other studies, etc.
How can we explain that the same types of bacteria are found in three different biotopes of HPW? One possible explanation is that the bacteria were contaminated during the sampling process or during preparation for examination. However, it is more likely that we are dealing with distinct strains of the same bacteria that may be unidentical in different biotopes [32]. However, colonization can also occur in the uterus, during passage through the genital tract and natural feeding. In addition, it has previously been shown that various Lactobacillus spp. are present in the vagina and bowel in diverse combinations and that their diversity depends on the TRI [33].
In this regard, it is important to understand the relationships between the various taxa defined in all biotopes. Our material shows that a reliable consistency exists, and it varies depending on the several co-occurrencies from TRI to TRI. It has been established that the co-occurrence between the bladder–vaginal and bladder–bowel biotopes is more pronounced than the relationships of the MLVs in the vaginal–bowel biotopes. Also, the links between aerobes are higher than those between anaerobic ones.
Our results enhance existing knowledge regarding the composition of the microbiota in three biotopes and demonstrate the dynamics and interrelated behaviour of the bladder, vaginal and bowel microbiota throughout pregnancy in first-time HPW. Biotopes are characterised by a stable dominance of certain aerobes and anaerobes. Furthermore, some of these taxa are marked by an unchanged IDFs in given biotopes.
Limitations. The strength of our work lies in the first-present study of the bladder microbiota jointly with the vaginal and bowel microbiota. However, we observe the limitations of our study being the small cohort of HPW, the insufficient breadth of species composition identification inherent in an extended culture-based study compared to gene sequencing approach.
Conclusion
The microbiota of the pregnant woman is essential for her health and foetal well-being. It is likely that our discovered microbiota composition at gestational ages observes a crucial role in its straightforward course. It is natural to assume that variations in microbiota composition from the studied one may be involved in the development of several pregnancy complications. Extended culture allows the detection and observation of these changes in microbial composition without resorting to the complex expensive investigative techniques available in all antenatal clinics worldwide. In this regard, it is essential to pay special attention to the evaluation of the urinary microbiome to prevent acute kidney and urinary tract infections. Consequently, it will be crucial in the future to determine which abnormalities in the microbial composition of the urinary tract convey a potential risk of pregnancy-related complications, such as acute pyelonephritis.
References
1. Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Bäckhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, Bäckhed F, Isolauri E, Salminen S, Ley RE. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150(3):470-480. DOI: 10.1016/j.cell.2012.07.008
2. Di Simone N, Santamaria Ortiz A, Specchia M, Tersigni C, Villa P, Gasbarrini A, Scambia G, D’Ippolito S. Recent Insights on the Maternal Microbiota: Impact on Pregnancy Outcomes. Front Immunol. 2020;11:528202. DOI: 10.3389/fimmu.2020.528202
3. Yang H, Guo R, Li S, Liang F, Tian C, Zhao X, Long Y, Liu F, Jiang M, Zhang Y, Ma J, Peng M, Zhang S, Ye W, Gan Q, Zeng F, Mao S, Liang Q, Ma X, Han M, Gao F, Yang R, Zhang C, Xiao L, Qin J, Li S, Zhu C. Systematic analysis of gut microbiota in pregnant women and its correlations with individual heterogeneity. NPJ Biofilms Microbiomes. 2020;6(1):32. DOI: 10.1038/s41522-020-00142-y
4. Liang X, Wang R, Luo H, Liao Y, Chen X, Xiao X, Li L. The interplay between the gut microbiota and metabolism during the third trimester of pregnancy. Front Microbiol. 2022;13:1059227. DOI: 10.3389/fmicb.2022.1059227
5. Haddad EN, Nel NH, Petrick LM, Kerver JM, Comstock SS. Associations between the Gut Microbiota, Urinary Metabolites, and Diet in Women during the Third Trimester of Pregnancy. Curr Dev Nutr. 2022;7(4):100025. DOI: 10.1016/j.cdnut.2022.100025
6. García-Mantrana I, Selma-Royo M, González S, Parra-Llorca A, Martínez-Costa C, Collado MC. Distinct maternal microbiota clusters are associated with diet during pregnancy: impact on neonatal microbiota and infant growth during the first 18 months of life. Gut Microbes. 2020;11(4):962-978. DOI: 10.1080/19490976.2020.1730294
7. Wang J, Zheng J, Shi W, Du N, Xu X, Zhang Y, Ji P, Zhang F, Jia Z, Wang Y, Zheng Z, Zhang H, Zhao F. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut. 2018;67(9):1614-1625. DOI: 10.1136/gutjnl-2018-315988
8. Zheng W, Xu Q, Huang W, Yan Q, Chen Y, Zhang L, Tian Z, Liu T, Yuan X, Liu C, Luo J, Guo C, Song W, Zhang L, Liang X, Qin H, Li G. Gestational Diabetes Mellitus Is Associated with Reduced Dynamics of Gut Microbiota during the First Half of Pregnancy. mSystems. 2020;5(2):e00109-20. DOI: 10.1128/mSystems.00109-20
9. Ma S, You Y, Huang L, Long S, Zhang J, Guo C, Zhang N, Wu X, Xiao Y, Tan H. Alterations in Gut Microbiota of Gestational Diabetes Patients During the First Trimester of Pregnancy. Front Cell Infect Microbiol. 2020;10:58. DOI: 10.3389/fcimb.2020.00058
10. Lv LJ, Li SH, Li SC, Zhong ZC, Duan HL, Tian C, Li H, He W, Chen MC, He TW, Wang YN, Zhou X, Yao L, Yin AH. Early-Onset Preeclampsia Is Associated With Gut Microbial Alterations in Antepartum and Postpartum Women. Front Cell Infect Microbiol. 2019;9:224. DOI: 10.3389/fcimb.2019.00224
11. Tersigni C, D’Ippolito S, Di Nicuolo F, Marana R, Valenza V, Masciullo V, Scaldaferri F, Malatacca F, de Waure C, Gasbarrini A, Scambia G, Di Simone N. Correction to: Recurrent pregnancy loss is associated to leaky gut: a novel pathogenic model of endometrium inflammation? J Transl Med. 2019;17(1):83. Erratum for: J Transl Med. 2018;16(1):102. DOI: 10.1186/s12967-019-1823-5.
12. D’Ippolito S, Tersigni C, Marana R, Di Nicuolo F, Gaglione R, Rossi ED, Castellani R, Scambia G, Di Simone N. Inflammosome in the human endometrium: further step in the evaluation of the “maternal side”. Fertil Steril. 2016;105(1):111-8.e1-4. DOI: 10.1016/j.fertnstert.2015.09.027
13. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108 Suppl 1(Suppl 1):4680-4687. DOI: 10.1073/pnas.1002611107
14. Anahtar MN, Gootenberg DB, Mitchell CM, Kwon DS. Cervicovaginal Microbiota and Reproductive Health: The Virtue of Simplicity. Cell Host Microbe. 2018;23(2):159-168. DOI: 10.1016/j.chom.2018.01.013
15. Hyman RW, Fukushima M, Jiang H, Fung E, Rand L, Johnson B, Vo KC, Caughey AB, Hilton JF, Davis RW, Giudice LC. Diversity of the vaginal microbiome correlates with preterm birth. Reprod Sci. 2014;21(1):32-40. DOI: 10.1177/1933719113488838
16. Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, Huang B, Arodz TJ, Edupuganti L, Glascock AL, Xu J, Jimenez NR, Vivadelli SC, Fong SS, Sheth NU, Jean S, Lee V, Bokhari YA, Lara AM, Mistry SD, Duckworth RA 3rd, Bradley SP, Koparde VN, Orenda XV, Milton SH, Rozycki SK, Matveyev AV, Wright ML, Huzurbazar SV, Jackson EM, Smirnova E, Korlach J, Tsai YC, Dickinson MR, Brooks JL, Drake JI, Chaffin DO, Sexton AL, Gravett MG, Rubens CE, Wijesooriya NR, Hendricks-Muñoz KD, Jefferson KK, Strauss JF 3rd, Buck GA. The vaginal microbiome and preterm birth. Nat Med. 2019;25(6):1012-1021. DOI: 10.1038/s41591-019-0450-2
17. Petrova MI, Lievens E, Malik S, Imholz N, Lebeer S. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front Physiol. 2015;6:81. DOI: 10.3389/fphys.2015.00081
18. Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, Strauss JF, The Vaginal Microbiome Consortium, Jefferson KK, Buck GA. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology (Reading). 2014;160(Pt 10):2272-2282. DOI: 10.1099/mic.0.081034-0
19. Abdelmaksoud AA, Koparde VN, Sheth NU, Serrano MG, Glascock AL, Fettweis JM, Strauss JF, Buck GA, Jefferson KK. Comparison of Lactobacillus crispatus isolates from Lactobacillus-dominated vaginal microbiomes with isolates from microbiomes containing bacterial vaginosis-associated bacteria. Microbiology (Reading). 2016;162(3):466-475. DOI: 10.1099/mic.0.000238
20. Stout MJ, Zhou Y, Wylie KM, Tarr PI, Macones GA, Tuuli MG. Early pregnancy vaginal microbiome trends and preterm birth. Am J Obstet Gynecol. 2017;217(3):356.e1-356.e18. DOI: 10.1016/j.ajog.2017.05.030
21. Hu F, Sun X, Su Y, Huang M. The Dynamic Changes in the Composition and Diversity of Vaginal Microbiota in Women of Different Pregnancy Periods. Microorganisms. 2023;11(11):2686. DOI: 10.3390/microorganisms11112686
22. Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, Raza S, Rosenbaum S, Van den Veyver I, Milosavljevic A, Gevers D, Huttenhower C, Petrosino J, Versalovic J. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7(6):e36466. DOI: 10.1371/journal.pone.0036466
23. Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, Galuppi M, Lamont RF, Chaemsaithong P, Miranda J, Chaiworapongsa T, Ravel J. Correction: The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014;2(1):10. Erratum for: Microbiome. 2014;2(1):4. DOI: 10.1186/2049-2618-2-10
24. MacIntyre DA, Chandiramani M, Lee YS, Kindinger L, Smith A, Angelopoulos N, Lehne B, Arulkumaran S, Brown R, Teoh TG, Holmes E, Nicoholson JK, Marchesi JR, Bennett PR. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep. 2015;5:8988. DOI: 10.1038/srep08988
25. Yeruva T, Rajkumar H, Donugama V. Vaginal lactobacilli profile in pregnant women with normal & abnormal vaginal flora. Indian J Med Res. 2017;146(4):534-540. DOI: 10.4103/ijmr.IJMR_774_16
26. Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, Bokulich NA, Song SJ, Hoashi M, Rivera-Vinas JI, Mendez K, Knight R, Clemente JC. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. 2016;22(3):250-253. DOI: 10.1038/nm.4039
27. Verma J, Bag S, Saha B, Kumar P, Ghosh TS, Dayal M, Senapati T, Mehra S, Dey P, Desigamani A, Kumar D, Rana P, Kumar B, Maiti TK, Sharma NC, Bhadra RK, Mutreja A, Nair GB, Ramamurthy T, Das B. Genomic plasticity associated with antimicrobial resistance in Vibrio cholerae. Proc Natl Acad Sci U S A. 2019;116(13):6226-6231. DOI: 10.1073/pnas.1900141116
28. Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. The MaSuRCA genome assembler. Bioinformatics. 2013;29(21):2669-2677. DOI: 10.1093/bioinformatics/btt476
29. Naser SM, Dawyndt P, Hoste B, Gevers D, Vandemeulebroecke K, Cleenwerck I, Vancanneyt M, Swings J. Identification of lactobacilli by pheS and rpoA gene sequence analyses. Int J Syst Evol Microbiol. 2007;57(Pt 12):2777-2789. DOI: 10.1099/ijs.0.64711-0
30. Mehta O, Ghosh TS, Kothidar A, Gowtham MR, Mitra R, Kshetrapal P, Wadhwa N, Thiruvengadam R; GARBH-Ini study group; Nair GB, Bhatnagar S, Das B. Vaginal Microbiome of Pregnant Indian Women: Insights into the Genome of Dominant Lactobacillus Species. Microb Ecol. 2020;80(2):487-499. DOI: 10.1007/s00248-020-01501-0
31. DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, Sun CL, Goltsman DS, Wong RJ, Shaw G, Stevenson DK, Holmes SP, Relman DA. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A. 2015;112(35):11060-11065. DOI: 10.1073/pnas.1502875112
32. Goltsman DSA, Sun CL, Proctor DM, DiGiulio DB, Robaczewska A, Thomas BC, Shaw GM, Stevenson DK, Holmes SP, Banfield JF, Relman DA. Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome. Genome Res. 2018;28(10):1467-1480. DOI: 10.1101/gr.236000.118
33. Dobrut A, Gosiewski T, Pabian W, Bodaszewska-Lubas M, Ochonska D, Bulanda M, Brzychczy-Wloch M. The dynamics of vaginal and rectal Lactobacillus spp. flora in subsequent trimesters of pregnancy in healthy Polish women, assessed using the Sanger sequencing method. BMC Pregnancy Childbirth. 2018;18(1):350. DOI: 10.1186/s12884-018-1987-7
34. Jacobs KM, Thomas-White KJ, Hilt EE, Wolfe AJ, Waters TP. Microorganisms Identified in the Maternal Bladder: Discovery of the Maternal Bladder Microbiota. AJP Rep. 2017;7(3):e188-e196. DOI: 10.1055/s-0037-1606860
35. Ollberding NJ, Völgyi E, Macaluso M, Kumar R, Morrow C, Tylavsky FA, Piyathilake CJ. Urinary Microbiota Associated with Preterm Birth: Results from the Conditions Affecting Neurocognitive Development and Learning in Early Childhood (CANDLE) Study. PLoS One. 2016;11(9):e0162302. DOI: 10.1371/journal.pone.0162302
36. Abbai NS, Reddy T, Ramjee G. Prevalent bacterial vaginosis infection – a risk factor for incident sexually transmitted infections in women in Durban, South Africa. Int J STD AIDS. 2016;27(14):1283-1288. DOI: 10.1177/0956462415616038
37. Abd El Aziz MA, Sharifipour F, Abedi P, Jahanfar S, Judge HM. Secnidazole for treatment of bacterial vaginosis: a systematic review. BMC Womens Health. 2019;19(1):121. DOI: 10.1186/s12905-019-0822-2
38. Aghaizu A, Reid F, Kerry S, Hay PE, Mallinson H, Jensen JS, Kerry S, Kerry S, Oakeshott P. Frequency and risk factors for incident and redetected Chlamydia trachomatis infection in sexually active, young, multi-ethnic women: a community based cohort study. Sex Transm Infect. 2014;90(7):524-528. DOI: 10.1136/sextrans-2014-051607
39. Ahmed A, Earl J, Retchless A, Hillier SL, Rabe LK, Cherpes TL, Powell E, Janto B, Eutsey R, Hiller NL, Boissy R, Dahlgren ME, Hall BG, Costerton JW, Post JC, Hu FZ, Ehrlich GD. Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J Bacteriol. 2012;194(15):3922-3937. DOI: 10.1128/JB.00056-12
40. Ahrens P, Andersen LO, Lilje B, Johannesen TB, Dahl EG, Baig S, Jensen JS, Falk L. Changes in the vaginal microbiota following antibiotic treatment for Mycoplasma genitalium, Chlamydia trachomatis and bacterial vaginosis. PLoS One. 2020;15(7):e0236036. DOI: 10.1371/journal.pone.0236036
41. Algburi A, Volski A, Chikindas ML. Natural antimicrobials subtilosin and lauramide arginine ethyl ester synergize with conventional antibiotics clindamycin and metronidazole against biofilms of Gardnerella vaginalis but not against biofilms of healthy vaginal lactobacilli. Pathog Dis. 2015;73(5):ftv018. DOI: 10.1093/femspd/ftv018
42. Naboka Yu.L., Gudima I.A., Kogan M.I., Chernitskaya M.L. Bacterial spectrum of the urine in young healthy women. Urologiia. 2010;(5):7-10. (In Russian) eLIBRARY ID: 15242297; EDN: MVOFTX
43. Kogan MI, Naboka YL, Ibishev KS, Gudima IA, Naber KG. Human urine is not sterile — shift of paradigm. Urol Int. 2015;94(4):445-452. DOI: 10.1159/000369631
44. Wolfe AJ, Brubaker L. “Sterile Urine” and the Presence of Bacteria. Eur Urol. 2015;68(2):173-174. DOI: 10.1016/j.eururo.2015.02.041
45. Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, Fitzgerald M, Mueller ER, Schreckenberger P, Dong Q, Nelson DE, Brubaker L. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol. 2012;50(4):1376-1383. DOI: 10.1128/JCM.05852-11
46. Hooton TM, Roberts PL, Cox ME, Stapleton AE. Voided midstream urine culture and acute cystitis in premenopausal women. N Engl J Med. 2013;369(20):1883-1891 DOI: 10.1056/NEJMoa1302186
47. Kogan M.I., Naboka Y.L., Gudima I.A., Vorovb’eva N.V. Asymptomatic bacteriuria in pregnant women — the normal condition of healthy women urine. Urologiia. 2022;(6):5-8. (In Russian). DOI: 10.18565/urology.2022.6.5-8
48. Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain inter-pretation. J Clin Microbiol. 1991;29(2):297-301. DOI: 10.1128/jcm.29.2.297-301.1991
49. Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, Mueller ER, Brubaker L, Gai X, Wolfe AJ, Schreckenberger PC. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52(3):871-876. DOI: 10.1128/JCM.02876-13
50. Guadamuro L, Dohrmann AB, Tebbe CC, Mayo B, Delgado S. Bacterial communities and metabolic activity of faecal cultures from equol producer and non-producer menopausal women under treatment with soy isoflavones. BMC Microbiol. 2017;17(1):93. DOI: 10.1186/s12866-017-1001-y
51. Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Bieda J, Chaemsaithong P, Miranda J, Chaiworapongsa T, Ravel J. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2:18. DOI: 10.1186/2049-2618-2-18
52. Callahan BJ, DiGiulio DB, Goltsman DSA, Sun CL, Costello EK, Jeganathan P, Biggio JR, Wong RJ, Druzin ML, Shaw GM, Stevenson DK, Holmes SP, Relman DA. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc Natl Acad Sci U S A. 2017;114(37):9966-9971. DOI: 10.1073/pnas.1705899114
About the Authors
Yu. L. NabokaRussian Federation
Yulia L. Naboka — Dr.Sc.(Med), Full Prof.
Rostov-on-Don
Competing Interests:
None
N. V. Vorobyeva
Russian Federation
Natalia V. Vorobyeva.
Rostov-on-Don
Competing Interests:
None
I. A. Gudima
Russian Federation
Irina A. Gudima — Dr.Sc.(Med), Assoc. Prof. (Docent).
Rostov-on-Don
Competing Interests:
None
T. G. Sinyavskaya
Russian Federation
Tatiana G. Sinyavskaya — Cand.Sc. (Econ), Assoc.Prof. (Docent).
Rostov-on-Don
Competing Interests:
None
R. S. Ismailov
Russian Federation
Ruslan S. Ismailov — Cand.Sc.(Med).
Rostov-on-Don
Competing Interests:
None
K. G. Naber
Germany
Kurt G. Naber — M.D., Ph.D., Assoc. Prof. of Urology.
Munich
Competing Interests:
None
M. I. Kogan
Russian Federation
Mikhail I. Kogan — Dr.Sc.(Med), Full Prof., Honored Scientist of the Russian Federation.
Rostov-on-Don
Competing Interests:
None
Review
For citations:
Naboka Yu.L., Vorobyeva N.V., Gudima I.A., Sinyavskaya T.G., Ismailov R.S., Naber K.G., Kogan M.I. Composition and dynamics of bladder, vaginal and bowel microbiota during three trimesters in healthy pregnant women. Urology Herald. 2024;12(5):12-23. https://doi.org/10.21886/2308-6424-2024-12-5-12-23













































